1p3hN is
a chain of M.
tuberculosis GroEs and the others are
chains of E.coli
GroES. They
have 37% sequence identity as shown below.
|
1
2
3
4
5
6
7
8
9
10
1234567890123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890 |
1p3hN
Others
|
AKVNIKPLEDKILVQANEAETTTASGLVIPDTAKEKPQEGTVVAVGPGRWDEDGEKRIPLDVAEGDTVIYSKYGGTEIKYNGEEYLILSARDVLAVVSK
|| || | | | || | | |
| | | | ||| || | ||
|| | || || |
| || |
MNIRPLHDRVIVKRKEVETKSAGGIVLTGSAAAKSTRGEVLAVGNGRILENGEVKPLDVKVGDIVIFNDGYGVKSEKIDNEEVLIMSESDILAIVEA
|
|
<= Base Loop
=>
<=R
=>
<=T=>
|
*"<=Base Loop=>"
, "<=R=>", and
"<=T=>" indicate the position of the base loop, the roof
loop, and the 79-83 turn respectively.
Although the structural variations for the base loop in E. coli GroES
are almost
negligible (whole protein Cα RMSDs 0.42 ± 0.13
Å), the corresponding DALI sequence alignments with M.
tuberculosis GroES show remarkable variation in this region. [1]
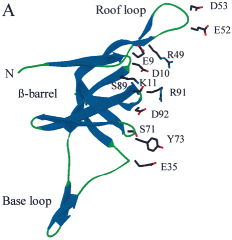
|
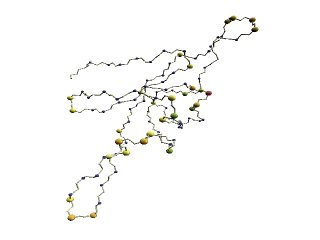
|
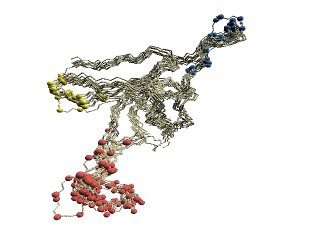 |
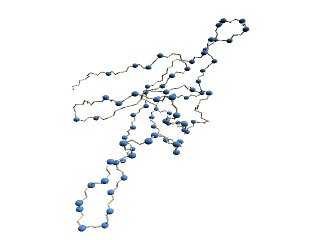
(A)
Structure of 1p3hN
|
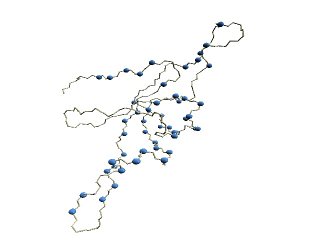
(B)
The D2 code of 1p3hN
|
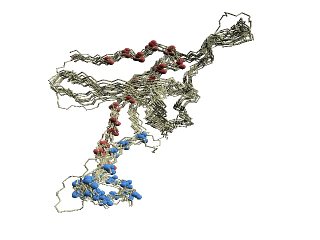
(C)
Structural variations of GroES. |
(A) Figure 5 of ref. [8].
(B) Residues are colored
according to their D
2 code: '0' in
blue, 'A' in red, 'B' and 'Q' in orange, 'G' in green, and 'R' in
yellow.
(C) The 'master-slave' alignments by DALI with 1p3hN as
master, where
residues of the base loop are shown in red, the roof loop in blue, and
the 79-83 turn in yellow.
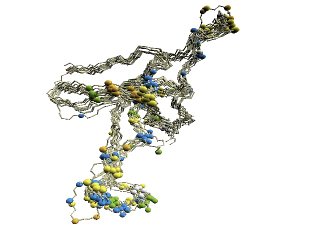 |
|
|
|
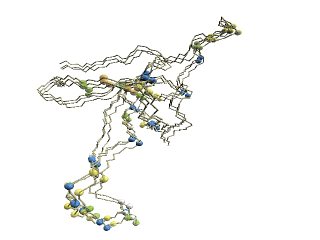
| (A)
D2 code-variable residues. |
(B)
HMM-SA
-variable residues. |
(C)
Stride-variable residues
|
(D)
Alignment blocks assigned by FATCAT
|
(A) The 'master-slave' alignments by
DALI
with 1p3hN as master, where residues with different
D2-codes
are colored accordingly: '0' in blue, 'A' in red, 'B' and 'Q'
in orange, 'G' in green, and 'R' in yellow.
(B) The structure of 1p3hN only is shown, where residues with different
HMM-SA
codes are shown in blue.
(C) The structure of 1p3h-N only is shown, where residues with
different
Stride
codes are shown in blue.
(D) Alignment blocks are colored as follows: residues in
block1 in red and
residues in
block2
in blue if multiple alignment blocks are assigned by
FATCAT.
The
D2 code of the
residues which do not have the same
D2 code as
the corresponding residue of 1p3hN is shown in the following figure.
Note that chains of 1pcq are assigned the same
D2 code-sequence. 1pf9O
and 1pf9R are also assigned the same
D2 code-sequence.
|
1
2
3
4
5
6
7
8
9
10
1234567890123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890 |
1p3hN
1aonO
1aonP
1aonS
1pf9O
1pf9R
1svtO
1pcqO
1pcqP
1pcqQ
1pcqR
1pcqS
|
--0000000R000000R00R000QBR0000RQBR0G000000R000R0000QBRB00O00RG00R000000RGR000000RR0000000QABG0000--
G
0 1G0R0RGR 0RR 0
RG
0 R RRG0 0
RG0
R0
R0QBG
G
G
0 1G1R0RGR 0RR 0
3G
0 R
RRG 0
RG0 0
R0QBG
G
0 1G1R0RGR 00R
0 G
0 R RRG0 0
RG0 R0 R0QBG
G
0 RG0R0RGRR00R G0
RG
0 R RRG0 0
RG0
0 0QBG
G
0 RG0R0RGRR00R G0
RG
0 R RRG0 0
RG0
0 0QBG
G 0 R RPG0R0RG 0RR0G0
RG
0 R1RR00 0
RG0
0O R0QBG
G
0 RPG0R0RG 0RR0 0 RG
0 R RR00 0
RG0
0O R0QBG
G 0
RPG0R0RG 0RR0 0
RG
0 R RR00 0
RG0
0O R0QBG
G 0
RPG0R0RG 0RR0 0
RG
0 R RR00 0
RG0
0O R0QBG
G 0
RPG0R0RG 0RR0 0
RG
0 R RR00 0
RG0
0O R0QBG
G 0
RPG0R0RG 0RR0 0
RG
0 R RR00 0
RG0
0O R0QBG
|
|
<= Base Loop
=>
<=R
=>
<=T=>
|
|
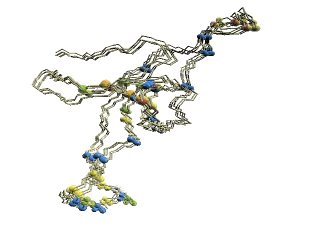
|
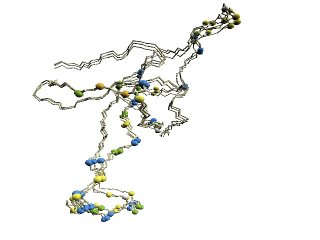 |
(A)
Chains of 1aon
|
(B) Chains
of 1pcq |
(C)
The other chains |
The 'master-slave' alignments by DALI with 1p3hN as master, where
residues with different D2-codes are colored accordingly: '0'
in
blue, 'A' in red, 'B' and 'Q' in orange, 'G' in green, and 'R' in
yellow. 1p3hN is not shown in the figures.
HMM-SA
(Hidden Markov Model-Structural Alphabet) is a collection of
27 structural prototypes of four residues. The
HMM-SA code of the
residues which do not have the same
HMM-SA code as
the corresponding residue of 1p3hN is shown in the following figure.
See below for the description of each alphabet.
|
1
2
3
4
5
6
7
8
9
10
1234567890123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890 |
1p3hN
1aonO
1aonP
1aonS
1pf9O
1pf9R
1svtO
1pcqO
1pcqP
1pcqQ
1pcqR
1pcqS
|
-SNMMNLJUSNLNMNGSKPRNMHaDSLMNHGBDOQKNLXKLGIMLHRJKLHaDEIMLLLPQTYUSLXMXKPOUSLLTTTUFSLXMMXKHBBQGILNT--
J L Y Q MXLLNJLU HESUFUQPRLYGFQMLGQ
LK
YPSUDOIKLLKPQMYUSLXMXKPSYBQX XL
QK
V LX
J L Y Q MX LNJLU KFSUFUQPRLYGFSN GQ
L
PSUDQJKL KPQMYCSLXLXKPSYBQXM L
K
EZ LX
L L Y Q X LNJLU HESUFUQPRNYGFSM GQ
L
PSUDQJKL KPQMYUSLXMXKPSYBQXMXL
K
EZ LX
L KY Q MX LNJLF HESURUQPRLYGFQM PQ
L
PSUDQJKN KPQNYUSLXMXKPSYBQT XL
K L
V LX
J KY Q MX LNJLF HESURUQPRLYGFQM PQ
L T PSUDQJKL
KPQNYUSLXMXKPSYBQT XL
K
V LX
L M Y QTMX LNJHFDDQGURUQLLLYGFQM GQ
L T PSUDSYJL
KPQNYCSLXMXKPSYBQN XM
K
PR
R Y QTMX LNJLFDDESURUQPRLYGFQM
GQ L T
PSUDORYLKKPQNYCSLXMXKPSYBQN
XL
LN
R Y QTMX LNJLFDDESURUQL LYGFQM
PQ L T
PSUDORYLKKPQNYCSLXMXKPSYBQN
XL
LN
R Y QTMX LNJLFDDESURUQPRLYGFQM
GQ L T
PSUDORYLKKPQNYCSLXMXKPSYBQN
X
LN
R Y QTMX LNJLFDDESURUQPRLYGFQM
GQ L T
PSUDORYLKKPQNYCSLXMXKPSYBQN
XL
LX
R Y QTMX LNJLFDDESURUQL LYGFQM
PQ L T
PSUDORYLKKPQNYCSLXMXKPSYBQN
XL
LN
|
|
<= Base Loop
=>
<=R
=>
<=T=>
|
STRIDE
[1] is a program to recognize secondary structural elements
in proteins from their atomic coordinates. It
performs the same task as
DSSP
by Kabsch and Sander but utilizes both
hydrogen bond energy and mainchain dihedral angles rather than
hydrogen bonds alone. See below for the description of each alphabet.
|
1
2
3
4
5
6
7
8
9
10
1234567890123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890 |
1p3hN
1aonO
1aonP
1aonS
1pf9O
1pf9R
1svtO
1pcqO
1pcqP
1pcqQ
1pcqR
1pcqS
|
EEEETTTEEEEEE BTTTT B
TTTT EEEEEEEEE EETTTT
EE TTTEEEEEETTTTEEEEETTEEEEEEEGGGEEEEEE
C
T
TTTTC TT C
T
CCB
CT C CC T
BCCBCCC
CCCCCCCC
CB
B
TTTTC TT C
T
CCB
CT C CC
T
CCBBTT
CCCCCBCC C
C T C
TTTTC TT C
T
CCBC
CT C CC
T
CCBCTT
CCCCCCCC C
C
T
TTTTT TT C
T
CCB
CT C CC T
BCCBBCC
CCCCC C C
C
T
TTTTT TT C
T
CCB
CT C CC T
BCCBCCC
CCCCCCC C
CCCB
B
TTTTC TT CCCC
CCB
E T
C
T
CCBCTT
CBTTTTC C
CCCB
B B TTTTT
TT CCCC CCBBCCC E
T
C
T
CCBCTT
CBTTTTC C
C
B B TTTTT
TT CCCC CCBBCCC E
T
C
T
CCBCTT
CBTTTTC C
C B CBB
TTTTT TT CCCC
CCBBCCC E T
C
T
CCBCTT
CBTTTTC C
C
B B TTTTT
TT CCCC CCBBCCC E
T
C
T
CCBCTT
CBCCCCC C
C
B
TTTTT TT CCCC
CCB
E T
C
T
CCBCTT
CBCCCCC C
|
|
<= Base Loop
=>
<=R
=>
<=T=>
|
Shown below is the 'master-slave' alignments by FATCAT with 1p3h-N as
master, where the numbers below amino acid-sequence indicate the
alignment block of the residue. Note that chains of 1pcq,
1pf9, and 1svt are decomposed into three alignment blocks
1,
2, and
3.
|
1
2
3
4
5
6
7
8
9
10
1234567890123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890 |
1p3hN
1aonO
1aonP
1aonS
1pf9O
1pf9R
1svtO
1pcqO
1pcqP
1pcqQ
1pcqR
1pcqS
|
VNIKPLEDKILVQANEA-ETTT----ASGLVIPDTAKEKPQEGTVVAVGPGRWDEDGEKRIPLDVAEGDTVIYSKYG-GTEIKYNGEEYLILSARDVLAVVSK
MNIRPLHDRVIVKRKEV-ETKSAGGIVLTGSAAAK----STRGEVLAVGNGRILE-NGEVKPLDVKVGDIVIFNDGYGVKSEKIDNEEVLIMSESDILAIVEA
11111111111111111-1111
111111111 1111111111111111
111111111111111111111 1111111111111111111111111
MNIRPLHDRVIVKRKEV-ETKSAGGIVLTGSAAAK----STRGEVLAVGNGRILE-NGEVKPLDVKVGDIVIFNDGYGVKSEKIDNEEVLIMSESDILAIVEA
11111111111111111-1111
111111111 1111111111111111
111111111111111111111 1111111111111111111111111
MNIRPLHDRVIVKRKEV-ETKSAGGIVLTGSAAAK----STRGEVLAVGNGRILE-NGEVKPLDVKVGDIVIFNDGYGVKSEKIDNEEVLIMSESDILAIVEA
11111111111111111-1111
111111111 1111111111111111
111111111111111111111 1111111111111111111111111
MNIRPLHDRVIVKRKEVETKSA----GGIVLTGSAAAKS-TRGEVLAVGNGRILE-NGEVKPLDVKVGDIVIFNDGYGVKSEKIDNEEVLIMSESDILAIVEA
11111111111111111
2222----2222222222222
333333333333333
33333333333333333333 33333333333333333333333333
MNIRPLHDRVIVKRKEVETKSA----GGIVLTGSAAAKS-TRGEVLAVGNGRILE-NGEVKPLDVKVGDIVIFNDGYGVKSEKIDNEEVLIMSESDILAIVEA
11111111111111111 2222----2222222222222
333333333333333
33333333333333333333 33333333333333333333333333
MNIRPLHDRVIVKRKEVETKSA----GGIVLTGSAAAKS-TRGEVLAVGNGRILE-NGEVKPLDVKVGDIVIFNDGYGVKSEKIDNEEVLIMSESDILAIVEA
11111111111111111 2222----2222222222222
333333333333333
33333333333333333333 33333333333333333333333333
MNIRPLHDRVIVKRKEVETKSA----GGIVLTGSAAAKS-TRGEVLAVGNGRILE-NGEVKPLDVKVGDIVIFNDGYGVKSEKIDNEEVLIMSESDILAIVEA
11111111111111111 2222----2222222222222
333333333333333
33333333333333333333 33333333333333333333333333
MNIRPLHDRVIVKRKEVETKSA----GGIVLTGSAAAKS-TRGEVLAVGNGRILE-NGEVKPLDVKVGDIVIFNDGYGVKSEKIDNEEVLIMSESDILAIVEA
11111111111111111 2222----2222222222222
333333333333333
33333333333333333333 33333333333333333333333333
MNIRPLHDRVIVKRKEVETKSA----GGIVLTGSAAAKS-TRGEVLAVGNGRILE-NGEVKPLDVKVGDIVIFNDGYGVKSEKIDNEEVLIMSESDILAIVEA
11111111111111111 2222----2222222222222
333333333333333
33333333333333333333 33333333333333333333333333
MNIRPLHDRVIVKRKEVETKSA----GGIVLTGSAAAKS-TRGEVLAVGNGRILE-NGEVKPLDVKVGDIVIFNDGYGVKSEKIDNEEVLIMSESDILAIVEA
11111111111111111 2222----2222222222222
333333333333333
33333333333333333333 33333333333333333333333333
MNIRPLHDRVIVKRKEVETKSA----GGIVLTGSAAAKS-TRGEVLAVGNGRILE-NGEVKPLDVKVGDIVIFNDGYGVKSEKIDNEEVLIMSESDILAIVEA
11111111111111111 2222----2222222222222
333333333333333
33333333333333333333 33333333333333333333333333
|
|
<= Base Loop
=>
<= R
=>
<=T=>
|









