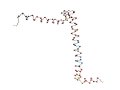
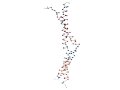
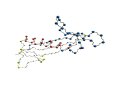
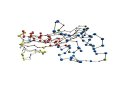
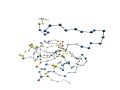
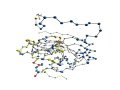
[Proteins with long variable regions]
- #lenN
stands for the number of occurrence of variable regions of length N.
- Backbone structures are obtained using ProteinViewer,
where
N-terminal is colored red and C-terminal blue.
- In the images, the CA atoms of the variable
regions are drawn as large
blue sphere.
- The "site"
row indicates active site residues:
"*" denotes active sites
registered
in SwissProt/UniProt.
"+" denotes active sites
registered in PROSITE/UniProt.
Residues
registered in both of them are denoted by "*."
- Click the images to view larger
image.
- See the bottom for brief explanation of
the DSSP
codes and the 5-tile
codes.
- "*" indicates the region of left- or
right-handed helical structure.
(1) Cluster 56: #len7 =
1, #len2 = 2
site
2beD
--RHQBG000000RQAAAAAAAAAAAJQAABGR0QAAAAAAAAA--
Res
LGDISGINASVVNIQKEIDRLNEVAKNLNESLIDLQELGKYEQYIK
2beF
--0RQBG000000RQAAAAAAAAAAAB0RRG000QAAAABQAAA--
|
| 5-tile
code sequences |
|
|
DSSP
LLLLLLLLLLLLLLHHHHHHHHHHHLLHhHHLLlhhhhhHHHHHHL......
2beD LGDISGINASVVNIQKEIDRLNEVAKNLnESLIdlqelgKYEQYIK......
||||||||||||||||||||||||||||
|
2beF LGDISGINASVVNIQKEIDRLNEVAKNL.NESL.....iDLQELGKyeqyik
DSSP
LLLLLLLLLLLLLLHHHHHHHHHHHHLL.LLLL.....lLHHHHLLhhhhhl
|
| Structural
alignment by DaliLite |
|
(2) Cluster 10: #len7 =
1, #len4 = 2, #len3 = 1, #len2 =
1, #len1 = 1
 |

|
|
No
Significant
Similarity
|
Z-Score: -
AR: -
RMSD: - A
SI:
- %
|
1d2z
C
res78-129
(len 52) |
1ik7
A
(len 52)
|
|
DaliLite result |
site
1d2z
--00QAAAAAAGAAB8BR00QAAAAAAAAABRRQAAAAABQABG0QAAAA--
Res
RGRSASNEFLNIWGGQYNHTVQTLFALFKKLKLHNAMRLIKDYVSEDLHKYI
1ik7
--AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA--
|
| 5-tile
code sequences |
|
|
DSSP
1d2z
1ik7
DSSP
|
| Structural
alignment by DaliLite |
|
(3) Cluster 39:
#len7 = 1, #len6 = 1, #len4 = 1, #len1 = 2
site
1mi7
--0QIAAAAAAAAAAAAAAAAAAAAAAAAGRRQAAAAAAB00QAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAABG0QAAAAAAAAA--
Res
QQSPYSAAMAEQRHQEWLRFVDLLKNAYQNDLHLPLLNLMLTPDEREALGTRVRIVEELLRGEMSQRELKNELGAGIATITRGSNSLKAAPVELRQWLEEVLL
1p6z
--R00QAAAAAAAAAAAAAAAAAAAAAABRRQAAAAAABG0QAAAAAAAAAAAAAAAAABRGG0QAAAAB0RR00QAAAAAAAAAAABG0QAAAAAAAAA--
|
| 5-tile
code sequences |
|
|
DSSP
LLLHhHHHHHHHHHHHHHHHHHHHHHHHLLLLHHHHHHHHLLHHHHHHHHHHHHHHHHHHHhhhhhhhhhhhhhhhhhhhhhhhhhhhhllhhhhhhhhhhhl
1mi7 QQSPySAAMAEQRHQEWLRFVDLLKNAYQNDLHLPLLNLMLTPDEREALGTRVRIVEELLRgemsqrelknelgagiatitrgsnslkaapvelrqwleevll
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||
1p6z QSPY.SAAMAEQRHQEWLRFVDLLKNAYQNDLHLPLLNLMLTPDEREALGTRVRIVEELLRgemsqrelknelgagiatitrgsnslkaapvelrqwleevll
DSSP
LLLL.LHHHHHHHHHHHHHHHHHHHHHHHHLLHHHHHHHHLLHHHHHHHHHHHHHHHHHHHllllhhhhhhhhlllhhhhhhhhhhhhhllhhhhhhhhhhhl
|
| Structural
alignment by DaliLite |
|
(4) Cluster 31: #len8
= 1, #len3 = 1, #len2 = 1, #len1 = 1 (, #len9 = 1)
site
2ou1
--RQAAAAAAAAAAAAAAAAAAAAAAAGQB01R1AAAAAGAAAAAAAAAAAAAABGQA---AAAAAAG1AA---
Res
AKEPCVESLVSQYFQTVTDYGKDLMEKVKSPELQAEAKSYFEKSKEQLTPLIKKAGTELVNFLSYFVELGTQPAQT
1l6l --00RHQAAABHQAAAAAAAAAAAAAAABHRGR0QAAAAAAAAAAAAAAAAAAAAABGQAAAAAG10000000--
|
| 5-tile
code sequences |
|
|
DSSP
....LLLL.HHHHhHHHHHHHHHHhHHHHHHHHhHLLLLLLHHHHHHHLLLLHHHHLHHHHLLL.LHHHHhhHHLL..ll
2ou1 ....EPCV.ESLVsQYFQTVTDYGkDLMEKVKSpELQAEAKSYFEKSKEQLTPLIKKAGTELVN.LSYFVelGTQP..at
|
|
|||||||||||||||||||||||||||||| |||||
1l6l akepCVESlVSQY.FQTVTDYGKD.LMEKVKSP.ELQAEAKSYFEKSKEQLTPLIKKAGTELVNfLSYFV..ELGTqpaq
DSSP
llllLLHHhHLLH.HHHHHHLLHH.HHHHHLLL.LLLHHHHHHHHHHHHHHHHHHHHHHLLHHHhHLLLL..LLLLllll
|
| Structural
alignment by DaliLite |
|
(5) Cluster 32:
#len11 = 1, #len6 = 1, #len3 = 1, #len2 = 1, #len1 = 1
site
2ou1
--RHRAAAAA-AAAAAAAAAAAAAAAAAB00RGQAAAAAAAAAAAAAAAAAAAAG1AAAAAAAAAAB1AA--
Res
EPCVSLVSQYFQTVTDYGKDLMEKVKSPELQAEAKSYFEKSKEQLTPLIKKAGTELVNFLSYFVELGTQPATQ
1l6l
--0PAAAAAAAAAAAAAAAAAAAAHB1QAAG0QBQAAAAAAAAA1AAAAAAAAAHQAAAAAAAAGG00000--
|
| 5-tile
code sequences |
|
|
DSSP
LLLL.LHHHHL.LHHHHHHHHHHHHHHLLLLLLlHHHHHHHLLLLHHHHHHLHHHHHHHHHHHHHhhHHLL...ll
2ou1 EPCV.SLVSQY.QTVTDYGKDLMEKVKSPELQAeAKSYFEKSKEQLTPLIKKAGTELVNFLSYFVelGTQP...at
||||
|||||||||||||||||||||||||||||||
1l6l EPCVsLVSQYFqTVTDYGKDLMEKVKSPELQAE.AKSYFEKSKEQLTPLIKKAGTELVNFLSYFV..ELGTqpatq
DSSP
LLLLlLHHHHHhHHHHHHHHHHHLLLLHHHHLL.LHHHHHHHHHHHHHHHHHLHHHHHHHHHHHL..LLLLlllll
|
| Structural
alignment by DaliLite |
|
(6) Cluster 52: #len11 =
1, #len6 = 1, #len3 = 1, #len1 = 4
site
1vrA
--00RGR000RG00R0000000000000QAAAAABRQBRG0000QB0RGR00QAQAAAAAAAAAAAAAAAABR0R000R000000000R000000000000--
Res
MIVTTTSGIQGKEIIEYIDIVNGEAIMGANIVRDLFASVRDVVGGRAGSYESKLKEARDIAMDEMKELAKQKGANAIVGVDVDYEVVRDGMLMVAVSGTAVRI
1vrD
--00RGR000RG00R0R0000000000000R00RG000QG0G0R00000QAAAAAAAAAAAAAAAAAAAAABR0R000R000000000RR00000000000--
|
| 5-tile
code sequences |
|
|
DSSP
LEELLLLLLLLLLLLEEEEEEEEEEEELhhhHHLLLH......hhlllLLLLllllLLLLHHHHHHHHHHHHHHHHHLLLLEEEEEEEEEEEEHHHEEEEEEEEEEEEL
1vrA MIVTTTSGIQGKEIIEYIDIVNGEAIMGaniVRDLFA......svrdvVGGRagsyESKLKEARDIAMDEMKELAKQKGANAIVGVDVDYEVVRDGMLMVAVSGTAVRI
||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||
1vrD MIVTTTSGIQGKEIIEYIDIVNGEAIMG...ANIVRDlfasvrdvvggRAGS...yESKLKEARDIAMDEMKELAKQKGANAIVGVDVDYEVVRDGMLMVAVSGTAVRI
DSSP
LEELLLLLLLLLLLLEEEEEEEEEEELL...LEEELLeelllllllllLLLL...hHHHHHHHHHHHHHHHHHHHHHLLLLEEEEEEEEEEEEHHHEEEEEEEEEEEEL
|
| Structural
alignment by DaliLite |
|
(7) Cluster 17: #len12 =
1, #len8 = 1, #len3 = 2, #len2 = 3, #len1 = 4
 |

|
|
NA
|
Z-Score:
26.9
AR: 199
RMSD: 2.0A
SI:
96%
|
| 1fm6
A (len 232) |
1g1u
A (len 229)
|
|
DaliLite result |
site
1fm6
--R00QAAAAAAAAB00000QABHQ00000000R00G2AAAAAAAAAAAAAAAAAAAABG0RQABG0QAAAAAAAAB1QAAAAAAAAAAA
Res
NEDMPVERILEAELAVEPKTETYVEANMGLNPSSPNDPVTNICQAADKQLFTLVEWAKRIPHFSELPLDDQVILLRAGWNELLIASFSHR
1g1u
--AAAAAAAAB0000QAAAB01RRQBBGR00000QAAAAAAAAAAAAAAAAAAABG0RQABG0QAAAAAAAAB1QAAAAAAAAAAA
site
1fm6
AQBG000000QBR00001AAAAABRR0QAAAAAA0B0QAAAAABR00QAAAAAAAAAAAB0QB00R0R0QAAAAAAAAAAAAAAAAAAAA
Res
SIAVKDGILLATGLHVHRNSAHSAGVGAIFDRVLTELVSKMRDMQMDKTELGCLRAIVLFNPDSKGLSNPAEVEALREKVYASLEAYCKH
1g1u
AQBG0G0000QBR0000QAAAAABRRQAAAAAAA0B0QAAAAABR00QAAAAAAAAAAAB0QBG0R0R0QAAAAAAAAAAAAAAAAAAAA
site
1fm6
B0QBGQBQAAAAAAAAAAAAAAAAAAAAAAAAAABB00R0001QAAAABR--
Res
KYPEQPGRFAKLLLRLPALRSIGLKCLEHLFFFKLIGDTPIDTFLMEMLEAPH
1g1u
B0QAGQBQAAAAAAAAAAAAAAAAAAAABRQAAABB1QAAAAAAAAAAABG--
|
| 5-tile
code sequences |
|
|
DSSP
llllLHHHHHHHHHLLLLLLLHHHH.lllLLLLLLLllllhhhhHHHHHHHHHHHHHHHHLLLLLLLLHHHHHHHHHHHHHHHHHHHHHHH
1fm6 nedmPVERILEAELAVEPKTETYVE.anmGLNPSSPndpvtnicQAADKQLFTLVEWAKRIPHFSELPLDDQVILLRAGWNELLIASFSHR
|||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||
1g1u ....PVERILEAELAVEPKTETYVEanmgLNPSSPN.dpvtnicQAADKQLFTLVEWAKRIPHFSELPLDDQVILLRAGWNELLIASFSHR
DSSP
....LHHHHHHHHHHLLLLLLLLLLllllLLLLLLL.lhhhhhhHHHHHHHHHHHHHHLLLLLHHHLLHHHHHHHHHHHHHHHHHHHHHHH
DSSP
LLLLLLEEELLLLLEEEHHHHHLLLLLLHHHHHHHHLHHHHHHLLLLHHHHHHHHHHHHLLLLLLLLLLHHHHHHHHHHHHHHHHHHHHH
1fm6
SIAVKDGILLATGLHVHRNSAHSAGVGAIFDRVLTELVSKMRDMQMDKTELGCLRAIVLFNPDSKGLSNPAEVEALREKVYASLEAYCKH
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
1g1u
SIAVKDGILLATGLHVHRNSAHSAGVGAIFDRVLTELVSKMRDMQMDKTELGCLRAIVLFNPDSKGLSNPAEVEALREKVYASLEAYCKH
DSSP
HHHLLLLEELLLLLEELHHHHHHLLLHHHHHHHHHHLHHHHHHHLLLHHHHHHHHHHHHLLLLLLLLLLHHHHHHHHHHHHHHHHHHHHH
DSSP
HLLLLLLHHHHHHLLHHHHHHHHHHHHHHH....hhhhllLLLLllllhhhhllll
1fm6
KYPEQPGRFAKLLLRLPALRSIGLKCLEHL....fffkliGDTPidtflmemleap
||||||||||||||||||||||||||||||
||
1g1u
KYPEQPGRFAKLLLRLPALRSIGLKCLEHLfffkligdtpIDTF...lmemleaph
DSSP
HLLLLLLHHHHHHLLHHHHHHHHHHHHHLLhhhhhhhhhhHHHH...hhhhhhlll
|
| Structural
alignment by DaliLite |
|
(8) Cluster 1: #len17 =
1, #len7 = 1, #len3 = 2, #len2 = 3, #len1 = 16
site
*
*
1a1q
--000000RG0G0000000QR10R1R0000RG00000QG000000000RG0000PGRGRGR0000QB000000000QBRRG0000000R000
Res
PITAYSQQTRGLLGCIITSLTGRDKNQVEGEVQVVSTATQSFLATCVNGVCWTVYHGAGSKTLAGPKGPITQMYTNVDQDLVGWQAPPGARS
1jxp
--00000R00QAAAAAAAABRG0RG0000OG00000R000000000RRG0000QAB00R00000R000000R000QABRG0003000R000
site
*
*
* * *
1a1q
00001RR0RG00000QBR000000000R0G000R0000QAABGRR0R00000QBR00R000000000RRG00000000QAAB0QABG00--
Res
LTPCTCGSSDLYLVTRHADVIPVRRRGDSRGSLLSPRPVSYLKGSSGGPLLCPSGHAVGIFRAAVCTRGVAKAVDFVPVESMETTMRSPVF
1jxp
0000RRR0RG00000QBR0R00000R0RRG000Q8R00QAAB0RR1RG0000QBR00000R000R00R0G0R000000QAAAAA--
|
| 5-tile
code sequences |
|
|
DSSP
llllllllllllllllllllllllLLLLLLLEEEEEELLEEEEEEELLLEEEEELLLLLLllLEELL..EELLLLEELLLLLEEEEELLLLLLL
1a1q pitaysqqtrgllgciitsltgrdKNQVEGEVQVVSTATQSFLATCVNGVCWTVYHGAGSktLAGPK..GPITQMYTNVDQDLVGWQAPPGARS
||||||||||||||||||||||||||||||||||||
|||||||||||||||||||||||||
1jxp .itaysqqtrgllgciitsltgrdKNQVEGEVQVVSTATQSFLATCVNGVCWTVYHGAGS..KTLAGpkGPITQMYTNVDQDLVGWQAPPGARS
DSSP
.llllllllllhhhhhhhhhllllLLLLLLLEEEEELLLLEEEEEEELLEEEEEHHHHLL..LLEEEllEEELLLEEELLLLEEEEELLLLLLL
DSSP
LLLLLLLLLEEEELLLLLLLEEEEEEELLEEEEEEEELHHHHLLLLLLEEELLLLLEEEEEEEEELLLLLEEEEEEEELLLLLLLLleell
1a1q
LTPCTCGSSDLYLVTRHADVIPVRRRGDSRGSLLSPRPVSYLKGSSGGPLLCPSGHAVGIFRAAVCTRGVAKAVDFVPVESMETTMrspvf
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
1jxp
LTPCTCGSSDLYLVTRHADVIPVRRRGDSRGSLLSPRPVSYLKGSSGGPLLCPSGHAVGIFRAAVCTRGVAKAVDFVPVESMETTM.....
DSSP
LLLLLLLLLEEEEELLLLLEEEEEELLLLEEEEEEEEEHHHHLLLLLLEEEELLLEEEEEEEEEEEELLEEEEEEEEEHHHHHLLL.....
|
| Structural
alignment by DaliLite |
|
[The DSSP code]
- H = alpha helix
- B = residue in isolated beta-bridge
- E = extended strand, participates in beta
ladder
- G = 3-helix (3/10 helix)
- I = 5 helix (pi helix)
- T = hydrogen bonded turn
- S = bend
- L = no assigned structure
[The 5-tile code]
- 0 = DDDDD (extended
strand, small
blue
spheres)
- 1 = DDDDU
- 2 = DDDUD
- 3 = DDDUU
- 8 = DUDDD
- 9 = DUDDU
- A = DUDUD (helix, large
red
spheres)
- B = DUDUU (C-cap, large
orange
spheres)
- G = UDDDD
- H = UDDDU
- I = UDDUD
- J = UDDUU
- O = UUDDD
- P = UUDDU
- Q = UUDUD (N-cap,
large orange spheres)
- R = UUDUU (turn,
large yellow spheres)