| 1 |
471
(60.8%)
|
|
AA:
V
Left: 0
Right: R
|
AA:
D
Left: 0
Right: G
|
AA:
I
Left: B
Right G
|
|
|
|
|
|
|
| 2 |
133
(17.2%)
|
|
AA:
QW
Left: 00
Right: RG
|

AA:
KY
Left: AA
Right: BQ
|
AA:
QT
Left: QB
Right R1
|
|
|
|
|
|
|
| 3 |
66
(8.5%)
|
|
AA:
RIV
Left: AAA
Right: B1Q
|
AA:
DPV
Left: 00Q
Right: G2A
|
AA:
YGS
Left: 0R0
Right RQB
|
|
|
|
|
|
|
| 4 |
49
(6.3%)
|
|
AA:
SQQS
Left: 0RRR
Right: R0QG
|
AA:
LKLH
Left: AAAA
Right: BRRQ
|

AA:
TGTN
Left: AAB0
Right BG0G
|
|
|
|
|
|
|
| 5 |
18
(2.3%)
|
|
AA:
EDDHH
Left: 0RQBO
Right: R0RRR
|
AA:
GEADC
Left: 00001
Right: QBQBG
|
AA:
KDTDS
Left: AAAAA
Right G0GRQ
|
|
|
|
|
|
|
| 6 |
15
(1.9%)
|
|
AA:
DIDATF
Left: 0QBRR0
Right: RG0QBG
|
AA:
RGEMSQ
Left: AAAAAA
Right: BRGG0Q
|
AA:
WFGNAQ
Left: AAAB00
Right B000RG
|
|
|
|
|
|
|
| 7 |
10
(1.3%)
|
|
AA:
NELGAGI
Left: AAAAAAA
Right: B0RR00Q
|
AA:
VADKGYT
Left: 00RRRRG
Right: R300000
|
AA:
YHGAGSK
Left: RGRGRGB
Right: QAB00R0
|
|
|
|
|
|
|
| 8 |
3
(0.4%)
|
|
AA:
DAVTFVNA
Left: 00000ABH
Right: 1BGRG0RQ
|
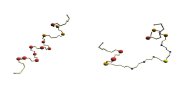
AA:
GDTPIDTF
Left: 1QAAAAAA
Right: 00R0001Q
|
AA:
KSPELQAE
Left: AGQB01R1
Right BHRGR0QA
|
|
|
|
|
|
|
| 9 |
2
(0.3%)
|
|
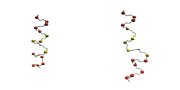
AA:
KTPETAGLI
Left: 00QB00RQG
Right: QG01RG00R
|
AA:
QHTIDLTDS
Left: AAAAAAAAA
Right: BRR00QBGQ
|
AA:
-
Left: -
Right -
|
|
|
|
|
|
|
| 10 |
1 (cluster 15)
(0.1%)
|
|
AA:
SSGAASTQSL
Left: 00000QBR00
Right: QB1RG000RG
|
AA:
-
Left: -
Right: -
|
AA:
-
Left: -
Right -
|
|
|
|
|
|
|
| 11 |
3
(0.4%)
|
|
AA:
VKSPELQAEAK
Left: AAAAB00RGQA
Right: HB1QAAG0QBQ
|
AA:
EGEETITAFVE
Left: 0RR0RBBQBAA
Right: RG0R01QAABQ
|
AA:
ANIVRDLFASV
Left: 00R00RG000Q
Right QAAAAABRQBR
|
|
|
|
|
|
|
| 12 |
1 (cluster 17)
(0.1%)
|
|
AA:
YVEANMGLNPSS
Left: AB01RRQBBGR0
Right: BHQ00000000R
|
AA:
-
Left: -
Right: -
|
AA:
-
Left: -
Right -
|
|
|
|
|
|
|
| 17 |
1 (cluster 01)
(0.1%)
|
|
AA:
RGLLGCIITSLTGRDKN
Left: 00QAAAAAAAABRG0RG
Right: G0G0000000QR10R1R
|
|
|
|
|
|
|
| 21 |
1 (cluster 22)
(0.1%)
|
|
AA:
IEKTNEKFHQIEKEFSEVEGR
Left: AAAAAAAAAAAAAAAAAAAAA
Right: BG0000000000000R00001
|
|
|
|
|
|
|
| 38 |
1 (cluster 48)
(0.1%)
|
|
AA:
LDMGNEIDTQNRQIDRIMEKADSNKTRIDEANQRATKM
Left: AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA
Right: B01RG01RRG0000000000RRR00000000000R00R
|
|
|
|
|
|
|

























