 |
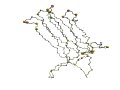 |
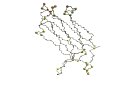
"The first beta-strand in 1rsy corresponds to the last beta-strand in 1qas and the topologies of the two domains are related by a circular permutation." (Grishin, 2001) | |
| 1rsy (Res142-262) | 1qas
(Res629-756) |
| 1rsy
<KLQYSLDYDFQNN
QLLVGIIQAAELPALDMGGTSDP YVKVFLLPDK K KKFETKVHRKTLN code <000000000QABR 00000R0000R000RGQBRG0R0 0000000O0R Q G00000000R0QG LCS 000000000 R 00000R0000R000 0 0 0000000 0 Q G00000000 0 G code R00000000000 R><300000R0000R000 0O010000000 0000QABG00000000 0RG 1qas PSATLFVKISIQ D><ERLRVRIISGQQLPK VNIVDPKVIVE IHGVGRDTGSRQTAVI TNN 1rsy PVFNEQFTFKVP YSELGGKTLVMAVYDFDRFSKHDIIGEFKVP MNTVDFGHVTEEWR> code 00OR00000R00 QAAAABG0000000000R000000R000000 QABG01RG000000> LCS 00 R00000 00 QA BG0000000000R000000 000000 QABG0 000000 code QB0000 R00000 00R0QA BG0000000000R000000 0000000QABG0 0000000000QB 1qas GFNPRW DMEFEF EVTVPD LALVRFMVEDYDSSSKNDF IGQSTIPWNSLK QGYRHVHLLSKN *RATIO : lcs/trimmed_seq_a = 0.803 (= 94/ 117), lcs/trimmed_seq_b = 0.790 (= 94/ 119) |
| Structural alignment by ComSubstruct |
| DSSP <...LLEEEEEEEEELll
LEEEEEEEEEELLLLLLlllLLLEEEEEEEEL..llLLLEELLLLLLLL.L 1rsy <...LGKLQYSLDYDFqn NQLLVGIIQAAELPALDmggTSDPYVKVFLLP..dkKKKFETKVHRKTL.N | | | || || || | | | | | 1qas qhpSATLFVKISIQD..><ERLRVRIISGQQLPKVN...IVDPKVIVEIHGvgrdTGSRQTAVITNNGfN DSSP lllLLEEEEEEEEEL..><LEEEEEEEEEELLLLLL...LLLEEEEEEEELllllLEEEELLLLLLLLlL DSSP LEEEEEEEELLLhhHHLLLEEEEEEEELLLLLLLEEEEEEEEEHHHLLlllLEEEEEEL......> 1rsy PVFNEQFTFKVPysELGGKTLVMAVYDFDRFSKHDIIGEFKVPMNTVDfghVTEEWRDL......> | | | | | | | || | || | | | 1qas PRWDMEFEFEVT..VPDLALVRFMVEDYDSSSKNDFIGQSTIPWNSLK...QGYRHVHLlskngd DSSP LEEEEEEEEELL..LHHHLEEEEEEEELLLLLLLEEEEEEEEEHHHLL...LEEEEEELllllll *Z-score 15.6 AR 109 RMSD 1.8A SI 27% |
| Structural alignment by DaliLite |

