 |
 |
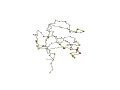
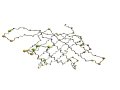
"Triabin shares significant and easily detectable sequence similarity with retinol-binding proteins and structurelly is more similar to them than any other protein family. However, the N-terminal regions of the structures (beta-strand abcd ...) are topologically distinct and thus triabin and retinol-binding protein can be classified into different folds." (Grishin, 2001) | |
| 1hbq (Res15-142) | 1avg
(ResI15-132) |
| 1hbq
KAR FAGTWYAMAKKDPEGL FLQDN
IVAEFSVDENGQMSAT AKGRVR
LLNNWDVCAD MVGTFTD TE DPA code QAA BG00000R000000R0 0001R 00000000QBR00000 000000 0QBR000000 0000000 0R GQB LCS QAA BG00000 0 R0 00 R 00000000 R 0 000000 0QB 0 0000000 0R G code QAAHBG00000 0 R0R00 RGO00000000 R 0RRRR000000R0QBG 0QBG0000000R0RRGR 1avg PEEFFNGTWYL A HGPGV TSPAVCQKFTT S GSKGFTQIVEIGYNKF ESNVKFQCNQVDNKNGE 1hbq KFKMKYWGVASFLQKGNDDHWIIDTD YETFAVQYSCRLLNLDGTCADSYSFVFA RD code G0000000RGQBG000000000R00R QG00000000R000QBRG0R00000000 RG LCS G0000000 QB 000000000 00 Q 00000000 000 RG0 00000000 RG code G0000000 QBR000000000 0000QB00000000 000 RG0 0000000000RG 1avg QYSFKCKS SDNTEFEADFTF ISVSYDNFALVCRS ITF TSQ PKEDRYLVFERT *RATIO : lcs/trimmed_seq_a = 0.742 (= 92/ 124), lcs/trimmed_seq_b = 0.807 (= 92/ 114) |
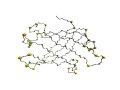
| Structural alignment by ComSubstruct |
| DSSP LLLLLL.LEEEEEEEeelllllllLEEEeeeeeellLLLEeeeeeeeeellllleEEEEEEEEEEE.LLLLl 1hbq FDKARF.AGTWYAMAkkdpeglflQDNIvaefsvdeNGQMsatakgrvrllnnwdVCADMVGTFTD.TEDPa | | |||| | 1avg FKPEEFfNGTWYLAH....gpgvtSPAV.cqkfttsGSKG..ftqiveigynkfeSNVKFQCNQVDnKNGE. DSSP LLHHHHlLLEEEEEL....lllllLLEE.eeeeellLLLL..leeeeeellllllLLLEEEEEELLlLLLL. DSSP EEEEEEEELLllLLLEEEEEEEEEELLLLEEEEEEEEeellllleEEEEEEEEELLLL 1hbq KFKMKYWGVAsfLQKGNDDHWIIDTDYETFAVQYSCRllnldgtcADSYSFVFARDPS | | | | || || | | 1avg QYSFKCKSSD..NTEFEADFTFISVSYDNFALVCRSI..tftsqpKEDRYLVFERTKS DSSP LEEEEEEELL..LLLEEEEEEEEEELLLLEEEEEEEE..eellllLLEEEEEEELLLL *Z-score 6.3 AR 85 RMSD 2.6A SI 20% |
| Structural alignment by DaliLite |