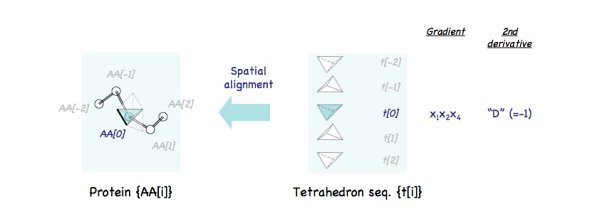
Align
therahedron t[0] with amino acid AA[0] and set the initial values.
The blue tetrahedron is aligned as shown on the left. And initial values, x1x2x4 and D, are assigned to t[0]. Note that "initial positions" of adjacent tiles t[1] and t[-1] are also determined (white tetrahedrons), which are moved to the position of AA[1] and AA[-1] respectively later.
The blue tetrahedron is aligned as shown on the left. And initial values, x1x2x4 and D, are assigned to t[0]. Note that "initial positions" of adjacent tiles t[1] and t[-1] are also determined (white tetrahedrons), which are moved to the position of AA[1] and AA[-1] respectively later.
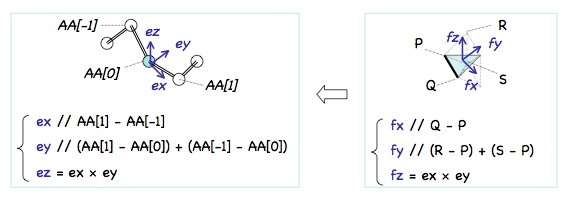
Orientation of AA[0] is specified by three unit vectors, ex, ey, and ez as shown on the left.
(In the figure, "A - B" denotes the vector from point B to A.)
Orientation of t[0] is specified by three unit vectors fx, fy, and fz as shown on the right.
(P, Q, R, and S are vertices of tetrahedrons.)
t[0] is aligned with AA[0] in such a way that the two coordinate systems (ex, ey, ez) and (fx, fy, fz) coincide.
Finally, in the "ProteinEncoder" program, the size of the tetrahedrons are determined such that the length of the shorter edge is about one fifth of the distance between the centers of AA[1] and AA[-1]: |Q - P| = |AA[1] - AA[-1]| / 5.
(Exactly speaking, the length of the shorter edge is qual to sqrt(6)/(5/3) of the average distance between successive atoms (i.e., N-CA and CA-C) of protein backbones. See module compute_3d_coordinates() in pe_encoding_suite.c.)